
Creates an Ensemble Model using Mean/Median Averaging
Source:R/ensemble_average.R
ensemble_average.RdCreates an Ensemble Model using Mean/Median Averaging
Usage
ensemble_average(object, type = c("mean", "median"))Details
The input to an ensemble_average() model is always a Modeltime Table,
which contains the models that you will ensemble.
Averaging Methods
The average method uses an un-weighted average using type of either:
"mean": Performs averaging usingmean(x, na.rm = TRUE)to aggregate each underlying models forecast at each timestamp"median": Performs averaging usingstats::median(x, na.rm = TRUE)to aggregate each underlying models forecast at each timestamp
Examples
# \donttest{
library(tidymodels)
#> ── Attaching packages ────────────────────────────────────── tidymodels 1.3.0 ──
#> ✔ broom 1.0.9 ✔ recipes 1.3.1
#> ✔ dials 1.4.1 ✔ rsample 1.3.1
#> ✔ dplyr 1.1.4 ✔ tibble 3.3.0
#> ✔ ggplot2 3.5.2 ✔ tidyr 1.3.1
#> ✔ infer 1.0.9 ✔ tune 2.0.0
#> ✔ modeldata 1.5.1 ✔ workflows 1.3.0
#> ✔ parsnip 1.3.3 ✔ workflowsets 1.1.1
#> ✔ purrr 1.1.0 ✔ yardstick 1.3.2
#> ── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
#> ✖ purrr::discard() masks scales::discard()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ recipes::step() masks stats::step()
library(modeltime)
library(modeltime.ensemble)
library(dplyr)
library(timetk)
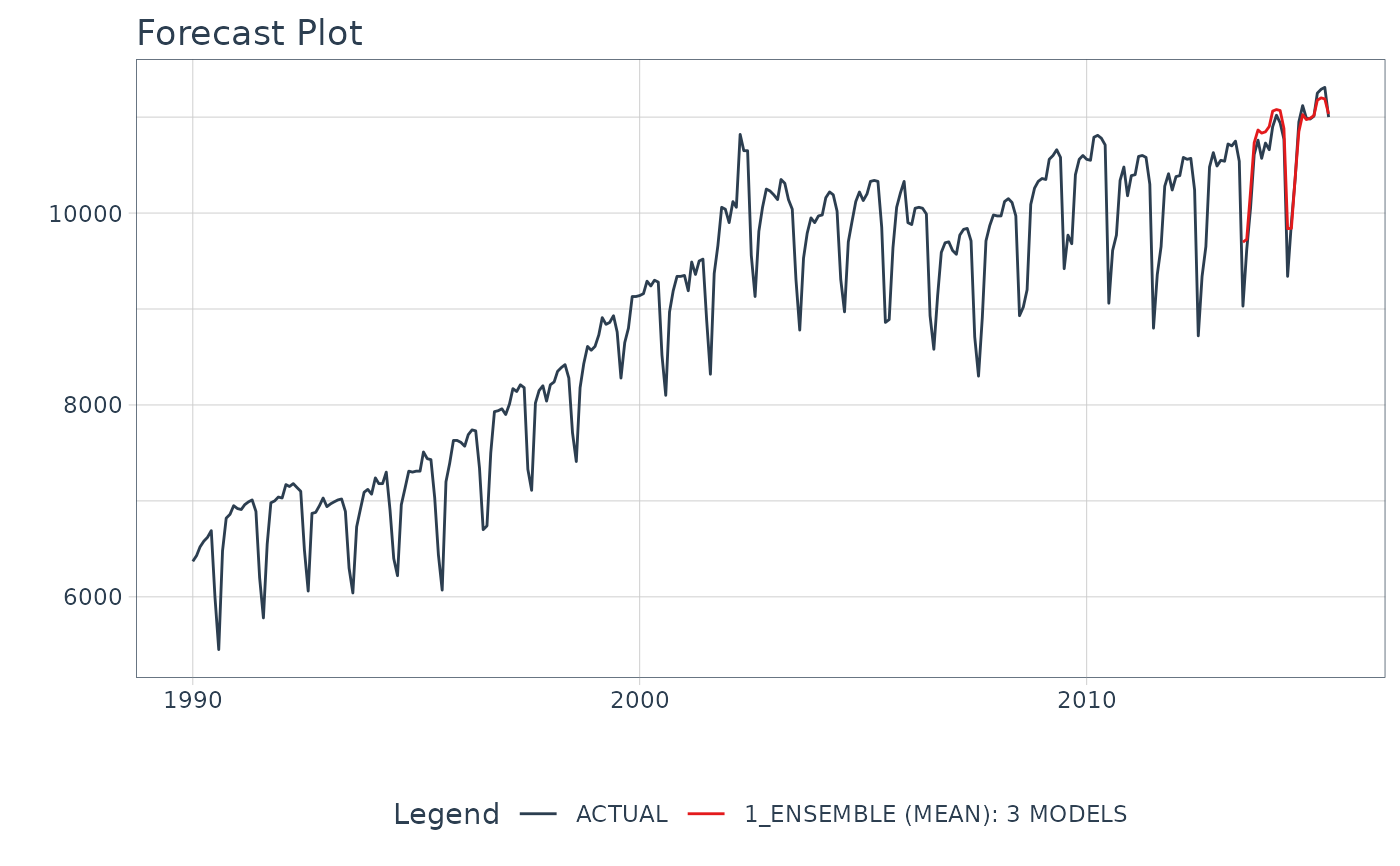
# Make an ensemble from a Modeltime Table
ensemble_fit <- m750_models %>%
ensemble_average(type = "mean")
ensemble_fit
#> ── Modeltime Ensemble ───────────────────────────────────────────
#> Ensemble of 3 Models (MEAN)
#>
#> # Modeltime Table
#> # A tibble: 3 × 3
#> .model_id .model .model_desc
#> <int> <list> <chr>
#> 1 1 <workflow> ARIMA(0,1,1)(0,1,1)[12]
#> 2 2 <workflow> PROPHET
#> 3 3 <workflow> GLMNET
# Forecast with the Ensemble
modeltime_table(
ensemble_fit
) %>%
modeltime_forecast(
new_data = testing(m750_splits),
actual_data = m750
) %>%
plot_modeltime_forecast(
.interactive = FALSE,
.conf_interval_show = FALSE
)
#> Warning: There were 2 warnings in `dplyr::mutate()`.
#> The first warning was:
#> ℹ In argument: `.nested.col = purrr::map2(...)`.
#> Caused by warning:
#> ! There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `.nested.col = purrr::map2(...)`.
#> Caused by warning:
#> ! `keep_original_cols` was added to `step_dummy()` after this recipe was created.
#> ℹ Regenerate your recipe to avoid this warning.
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 1 remaining warning.
 # }
# }